Identification of a new B.1.1.33 SARS-CoV-2 Variant of Interest (VOI) circulating in Brazil with mutation E484K and multiple deletions in the amino (N)-terminal domain of the Spike protein
Paola Cristina Resende 1a; Tiago Gräf 2a; Lidio Gonçalves Lima Neto 3, Fabiano Vieira da Silva 3, Anna Carolina Dias Paixão 1; Luciana Appolinario 1; Renata Serrano Lopes 1; Ana Carolina da Fonseca Mendonça 1; Alice Sampaio Barreto da Rocha 1; Fernando Couto Motta 1; Edson Delatorre 4** ; Gabriel L Wallau 5**; Felipe G Naveca 6**; Gonzalo Bello 7**, Marilda Mendonça Siqueira 1** on behalf of Fiocruz COVID-19 Genomic Surveillance Network
a These authors share the first authorship. ** These authors share the senior authorship
1. Laboratory of Respiratory Viruses and Measles (LVRS), Oswaldo Cruz Institute, Fiocruz
2. Plataforma de Vigilância Molecular, Instituto Gonçalo Moniz, Fiocruz-BA
3. Laboratório Central de Saúde Pública do Estado do Maranhão (LACEN-MA)
4. Departamento de Biologia. Centro de Ciências Exatas, Naturais e da Saúde, Universidade Federal do Espírito Santo, Alegre, Brazil.
5. Departamento de Entomologia e Núcleo de bioinformática, Instituto Aggeu Magalhães, Fiocruz-PE
6. Laboratório de Ecologia de Doenças Transmissíveis na Amazônia (EDTA), Leônidas e Maria Deane Institute, Fiocruz
7. Laboratório de AIDS e Imunologia Molecular, Oswaldo Cruz Institute, Fiocruz.
Summary
The SARS-CoV-2 2021 epidemic in Brazil is dominated by different lineages that harbor the E484K mutation at the receptor-binding domain (RBD) of the Spike protein (S). One lineage designated as P.1 is considered a Variant of Concern (VOC), while two other lineages designated as P.2 and N.9 are considered Variants of Interest (VOI). Here, we report a new SARS-CoV-2 VOI derived from lineage B.1.1.33, preliminary designated as PANGO lineage N.10, harboring 14 lineage defining mutations. It displays as the most remarkable genetic changes the V445A and E484K mutations in the S protein RBD, several non-synonymous mutations (P9L, I210V, and L212I), and three deletions (∆141-144, ∆211, and ∆256-258) in the S protein NTD, including a truncated NS7b protein due to a frame-shifting deletion. Other five lineage-defining mutations were found among NSP3, NSP5, NSP6, and N. This VOI probably emerged in late December 2020 and comprises a significant fraction (23%) of the SARS-CoV-2 positive cases detected in the Brazilian state of Maranhão (Northeastern region), between January and February 2021.
Background
Several SARS-CoV-2 variants with the mutation E484K at the receptor-binding site (RBD) of the Spike (S) protein have emerged in Brazil and dominate the epidemic since late 2020 (https://www.genomahcov.fiocruz.br). The Variant of Concern (VOC) P.1 and the Variant of Interest (VOI) P.2 have evolved from lineage B.1.1.28 (1-4), while the VOI N.9 evolved from lineage B.1.1.33 (5). In a previous genomic survey conducted by our group, we identified two B.1.1.33 sequences sampled in the Brazilian state of Maranhão (Northeast region) that harbors mutation E484K in the S protein, but branched outside the lineage N.9 (6). To determine whether these sequences were part of a new emergent SARS-CoV-2 lineage, herein we further sampled and analyzed the genetic diversity of SARS-CoV-2 viruses circulating in Maranhão in 2021.
The Study
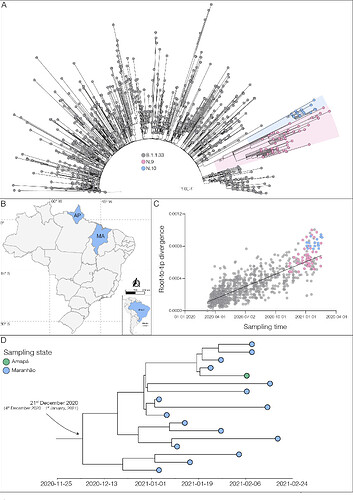
The Fiocruz COVID-19 Genomic Surveillance Network sequenced 73 SARS-CoV-2 positive samples (Appendix 1) detected in the state of Maranhão between 1st January and 24th February 2021. The mutational profile of the 73 SARS-CoV-2 genomes was investigated using the Nextclade tool (https://clades.nextstrain.org) and identified 70 sequences (96%) carrying the S:E484K amino acid change belonging to four different PANGO lineages (7): P.1 (n = 26), P.2 (n = 15), N.9 (n = 13) and B.1.1.33 (n = 16). The B.1.1.33 and N.9 sequences from the state of Maranhão here obtained were next combined with 891 B.1.1.33 and 38 N.9 high-quality complete genomes of Brazilian origin available in the EpiCoV database in GISAID by 31st March 2021 (Appendix Table 1). Maximum-likelihood (ML) phylogenetic analysis conducted using IQ-TREE v2.1.2 (8) revealed that all 16 B.1.1.33(E484K) sequences from Maranhão branched in a highly supported (approximate likelihood-ratio test [aLRT] = 100%) monophyletic clade, preliminary designated as PANGO lineage N.10 (Proposal for lineage within B.1.1.33 (Suggestion lineage N.10) · Issue #40 · cov-lineages/pango-designation · GitHub), that also comprises one sequence sampled in the state of Amapá (North region) in February 2021 (Figures 1a and 1b). Analysis of the temporal structure using TempEst (9) revealed that the overall divergence of lineage N.10 is above the mean of other B.1.1.33 and N.9 sequences (Figure 1c). Bayesian reconstruction using a strict molecular clock model with a uniform substitution rate prior (8 - 10 × 10-4 substitutions/site/year) as implemented in BEAST 1.10 (10) estimated the emergence of the VOI N.10 on 21st December 2020 (95% High Posterior Density [HPD]: 4th December 2020 – 1st January 2021) (Figure 1d).
Figure 1. a) Maximum likelihood (ML) phylogenetic tree of the B.1.1.33 diversity in Brazil. The VOIs N.9 and N.10 clades are highlighted in pink and blue, respectively. The aLRT support values are indicated in key nodes, and branch lengths are drawn to scale with the bar indicating nucleotide substitutions per site. b) Geographic distribution of the VOI N.10 identified in Brazil. MA - Maranhão state; AP - Amapá state, following the ISO 3166-2 standard. c) Correlation between the sampling date of B.1.1.33, N.9, and N.10 sequences and their genetic distance from the ML phylogenetic tree’s root. Colors represent N.9 and N.10 as indicated in a). d) Time-scaled maximum clade credibility tree estimated under a strict molecular clock’s assumption.
This new VOI N.10 spreading in Maranhão is characterized by 14 lineage-defining genetic changes, including ten non-synonymous mutations, three in-frame deletions, and one frame-shifting 4nt deletion (Table 1). Eight lineage-defining genetic changes were located in the S protein being two non-synonymous mutations at the RBD (E484K and V445A) and two non-synonymous mutations (I210V and L212I) and three deletions (∆141-144, ∆211 and ∆256-258) in the amino (N)-terminal domain (NTD).
Table 1. Synapomorphic mutations of SARS-CoV-2 lineage N.10.
| Genomic region (protein) | Nucleotide | Amino acid |
|---|---|---|
| ORF1ab (NSP3) | C5184T | P822L |
| ORF1ab (NSP5) | C10376T | P108S |
| C10478T | P132S | |
| ORF1ab (NSP6) | T11418C | V149A |
| Spike (S) | C21588T | P9L |
| ∆21984 - 21996 | ∆141-144 | |
| A22190G | I210V | |
| ∆22193 - 22195 | ∆211 | |
| T22196A | L212I | |
| ∆22327 - 22336 | ∆256-258 | |
| T22896C | V445A | |
| G23012A | E484K | |
| ORF7b (NSP7b) | ∆27794 - 27798 | frame-shifted and truncated |
| N | A28482G | Q70R |
The new VOI N.10 has several lineage-defining genetic changes with phenotypic implications. Mutation S:E484K has been identified as one of the most critical substitutions that could contribute to immune evasion (11-13). Mutation S:V445A is located in a loop (sites 443–450) in the receptor-binding motif (RBM, a portion of RBD making direct contact with ACE2) that is targeted by many anti-RBD neutralizing antibodies and mutations at that site might also confer partial resistance to neutralization (14). The NTD deletions ∆141-144, ∆211, and ∆256-258 are located within or close to the recurrent deletion regions (RDR) designated as RDR2 (139-146), RDR3 (210-212), and RDR4 (242-248). The RDR2 and RDR4 compose the NTD antigenic-supersite, and deletions at these regions confer resistance to neutralization by anti-NTD neutralizing antibodies (6, 15, 16). Moreover, mutation Q70R is located in a SARS-CoV-2-specific T cell epitope from Nucleocapsid (sites 66 to 74) (17). Finally, the 4nt deletion in the ORF7b leads to a truncated NS7b protein which resembles the Δ382 variant described in Singapore in early 2020 (18). Of note, this strain was associated with milder clinical outcomes; however, this cluster was extinct after successful control measures implemented by the Singapore government (19).
Taken together, these findings revealed that a new VOI N.10 carrying multiple mutations with phenotypic implications evolved within lineage B.1.1.33 and is currently spreading in the Northeastern Brazilian states of Maranhao. This new VOI could be partially resistant to anti-RBD and anti-NTD neutralizing antibodies and might also be associated with different clinical outcomes. These features make N.10 a variant whose dissemination should be closely monitored.
Acknowledgements
This study was approved by the FIOCRUZ-IOC Ethics Committee (CAAE: 68118417.6.0000.5248 and 32333120.4.0000.5190) and the Brazilian Ministry of the Environment (MMA) SISGEN (A1767C3). The authors wish to thank all the health care workers and scientists who have worked hard to deal with this pandemic threat, the GISAID team, and all the EpiCoV database’s submitters, GISAID acknowledgment table containing sequences used in this study is attached to this post (Appendix Table 1). We also appreciate the support of the Fiocruz COVID-19 Genomic Surveillance Network (http://www.genomahcov.fiocruz.br/) members, the Respiratory Viruses Genomic Surveillance. General Coordination of the Laboratory Network (CGLab), Brazilian Ministry of Health (MoH), Brazilian States Central Laboratories (LACENs), Brazilian Ministry of Health (MoH), and the Amazonas surveillance teams for the partnership in the viral surveillance in Brazil. Funding support FAPEAM (PCTI-EmergeSaude/AM call 005/2020 and Rede Genômica de Vigilância em Saúde - REGESAM); Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant 403276/2020-9); Inova Fiocruz/Fundação Oswaldo Cruz (Grant VPPCB-007-FIO-18-2-30 - Geração de conhecimento).
References:
-
Fujino T, Nomoto H, Kutsuna S, Ujiie M, Suzuki T, Sato R, et al. Novel SARS-CoV-2 Variant Identified in Travelers from Brazil to Japan. Emerg Infect Dis. 2021 Feb 10;27(4).
-
Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, et al. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new Variant of Concern P.1. Research Square; 2021.
-
Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DdS, Mishra S, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 2021-01-01 00:00:00.
-
Voloch CM, da Silva Francisco R, Jr., de Almeida LGP, Cardoso CC, Brustolini OJ, Gerber AL, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol. 2021 Mar 1.
-
Resende PC, Gräf T, Paixão ACD, Appolinario L, Lopes RS, da Fonseca Mendonça AC, et al. A potential SARS-CoV-2 variant of interest (VOI) harboring mutation E484K in the Spike protein was identified within lineage B.1.1.33 circulating in Brazil. bioRxiv. 2021 2021-01-01 00:00:00.
-
Resende PC, Naveca FG, Lins RD, Dezordi FZ, Ferraz MVF, Moreira EG, et al. The ongoing evolution of variants of concern and interest of SARS-CoV-2 in Brazil revealed by convergent indels in the amino (N)-terminal domain of the Spike protein. medRxiv. 2021 2021-01-01 00:00:00.
-
Rambaut A, Holmes EC, O’Toole A, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020 Jul 15.
-
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020 May 1;37(5):1530-4.
-
Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016 Jan;2(1):vew007.
-
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018 Jan;4(1):vey016.
-
Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020 Aug 21;369(6506):1014-8.
-
Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021 Feb 8.
-
Wang P, Liu L, Iketani S, Luo Y, Guo Y, Wang M, et al. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv. 2021 2021-01-01 00:00:00.
-
Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020 Oct 28;9.
-
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021 Mar 12.
-
Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020 Aug 7;369(6504):650-5.
-
Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021 Jan;22(1):74-85.
-
Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, et al. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio. 2020 Jul 21;11(4).
-
Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020 Aug 29;396(10251):603-11.
Appendix 1. GISAID accession numbers of 73 genomes from Maranhão produced by the Fiocruz COVID-19 Genomic Surveillance Network.
EPI_ISL_1181370, EPI_ISL_1181371, EPI_ISL_1181400, EPI_ISL_1181401, EPI_ISL_1181413, EPI_ISL_1181414, EPI_ISL_1465185, EPI_ISL_1465186, EPI_ISL_1465188, EPI_ISL_1465189, EPI_ISL_1465191, EPI_ISL_1465192, EPI_ISL_1465194 to EPI_ISL_1465196, EPI_ISL_1465198 to EPI_ISL_1465203, EPI_ISL_1465205, EPI_ISL_1465206, EPI_ISL_1465208 to EPI_ISL_1465210, EPI_ISL_1465212, EPI_ISL_1465213, EPI_ISL_1465215 to EPI_ISL_1465217, EPI_ISL_1465219 to EPI_ISL_1465222, EPI_ISL_1465224 to EPI_ISL_1465226, EPI_ISL_1465228, EPI_ISL_1465229, EPI_ISL_1465231, EPI_ISL_1465232, EPI_ISL_1465234 to EPI_ISL_1465236, EPI_ISL_1465238, EPI_ISL_1465239, EPI_ISL_1465241 to EPI_ISL_1465243, EPI_ISL_1465245, EPI_ISL_1465246, EPI_ISL_1465248 to EPI_ISL_1465250, EPI_ISL_1465252 to EPI_ISL_1465254, EPI_ISL_1465255, EPI_ISL_1465257, EPI_ISL_1465258, EPI_ISL_1465259, EPI_ISL_1465261, EPI_ISL_1465262, EPI_ISL_1465264, EPI_ISL_1465265, EPI_ISL_1465267, EPI_ISL_1465268, EPI_ISL_1465270, EPI_ISL_1465271, EPI_ISL_1465273 to EPI_ISL_1465275, EPI_ISL_1465277, EPI_ISL_1465278.
Appendix Table 1. GISAID acknowledgment table.gisaid_hcov-19_acknowledgement_table_2021_03_30_13.pdf (3.6 KB)