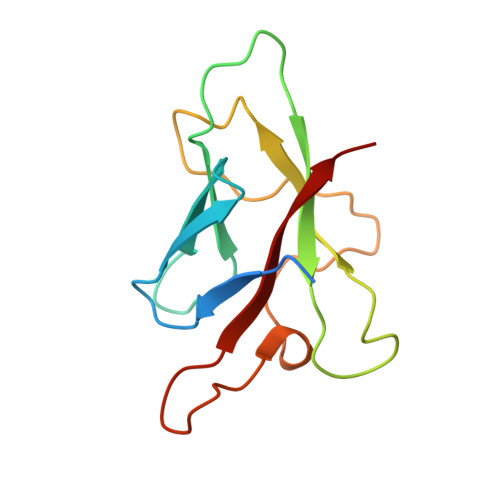
The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein.
Guncar, G., Wang, C.I., Forwood, J.K., Teh, T., Catanzariti, A.M., Ellis, J.G., Dodds, P.N., Kobe, B.(2007) Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 209-213
- PubMed: 17329816
- DOI: https://doi.org/10.1107/S1744309107004599
- Primary Citation of Related Structures:
2OPC - PubMed Abstract:
Metal-binding sites are ubiquitous in proteins and can be readily utilized for phasing. It is shown that a protein crystal structure can be solved using single-wavelength anomalous diffraction based on the anomalous signal of a cobalt ion measured on a conventional monochromatic X-ray source. The unique absorption edge of cobalt (1.61 A) is compatible with the Cu K alpha wavelength (1.54 A) commonly available in macromolecular crystallography laboratories. This approach was applied to the determination of the structure of Melampsora lini avirulence protein AvrL567-A, a protein with a novel fold from the fungal pathogen flax rust that induces plant disease resistance in flax plants. This approach using cobalt ions may be applicable to all cobalt-binding proteins and may be advantageous when synchrotron radiation is not readily available.
Organizational Affiliation:
School of Molecular and Microbial Sciences, University of Queensland, Brisbane, Qld 4072, Australia.